Supported DL frameworks
Pablo Morala
2024-01-17
Source:vignettes/nn2poly-02-supported-DL-frameworks.Rmd
nn2poly-02-supported-DL-frameworks.RmdIn this vignette we will explore the supported deep learning
frameworks in nn2poly and how can they be used. Currently
supported frameworks are keras/tensorflow and
luz/torch. The benefit of using them is that
we provide an easy to use implementation of the weight constraints
needed during training to avoid problems in the Taylor expansion carried
out by nn2poly. Furthermore, models created with these
frameworks are directly supported by the nn2poly()
function, allowing then to skip the need to manually create an object
with weights and activation functions as explained in
vignette("nn2poly-01-introduction").
If you are interested in having support for another deep learning framework, please open an issue in our GitHub repo!
Note: Using the provided constraints when training a model increases the number of needed epochs to converge, which is an expected trade off between learning speed and control over the weights for easier explainability.
Data generation
As our goal here is to show how to impose the needed constraints for
each framework, we will replicate the polynomial data generation from
vignette("nn2poly-01-introduction") and solve later the
same regression problem with the different deep learning frameworks.
Refer to that vignette for more details.
Create the polynomial :
Generate the data:
# Define number of variables and sample size
p <- 3
n_sample <- 500
# Predictor variables
X <- matrix(0,n_sample,p)
for (i in 1:p){
X[,i] <- rnorm(n = n_sample,0,1)
}
# Response variable + small error term
Y <- nn2poly:::eval_poly(poly = polynomial, newdata = X) +
stats::rnorm(n_sample, 0, 0.1)
# Store all as a data frame
data <- as.data.frame(cbind(X, Y))
head(data)
#> V1 V2 V3 Y
#> 1 1.3709584 1.029140719 2.3250585 -1.7547416
#> 2 -0.5646982 0.914774868 0.5241222 -3.7107357
#> 3 0.3631284 -0.002456267 0.9707334 1.3609395
#> 4 0.6328626 0.136009552 0.3769734 2.4608270
#> 5 0.4042683 -0.720153545 -0.9959334 -0.6141076
#> 6 -0.1061245 -0.198124330 -0.5974829 -0.7455793Scale the data to have everything in the interval and divide it in train and test.
# Data scaling to [-1,1]
maxs <- apply(data, 2, max)
mins <- apply(data, 2, min)
data <- as.data.frame(scale(data, center = mins + (maxs - mins) / 2, scale = (maxs - mins) / 2))
# Divide in train (0.75) and test (0.25)
index <- sample(1:nrow(data), round(0.75 * nrow(data)))
train <- data[index, ]
test <- data[-index, ]
train_x <- as.matrix(train[,-(p+1)])
train_y <- as.matrix(train[,(p+1)])
test_x <- as.matrix(test[,-(p+1)])
test_y <- as.matrix(test[,(p+1)])Supported frameworks examples
With our common data generated, we are now ready to delve into the
different deep learning frameworks. In each case we will solve the same
regression problem with the previously generated polynomial data, with
two neural networks for each framework. These neural networks will have
the same structure but one will be constrained (nn_con) and
the other one unconstrained (nn_uncon) to showcase their
differences.
keras/tensorflow
In this section we will show how to use nn2poly with a
keras neural network and how to impose the needed weight
constraints during training.
library(nn2poly)
library(keras)
# This sets all needed seeds
tensorflow::set_random_seed(42)Model definition
First of all we will set the structure of our neural networks using a
sequential model in keras. This following function will be
used to create both the constrained and unconstrained neural networks in
the keras example.
keras_model <- function() {
tensorflow::set_random_seed(42)
nn <- keras::keras_model_sequential()
nn <- keras::layer_dense(nn, units = 100, activation = "tanh", input_shape = p)
nn <- keras::layer_dense(nn, units = 100, activation = "tanh")
nn <- keras::layer_dense(nn, units = 100, activation = "tanh")
nn <- keras::layer_dense(nn, units = 1, activation = "linear")
nn
}Now we define both NNs. The only needed step to impose our
implemented weight constraints on a keras model is to use
the function add_constraints() and then train it as
usually. The constraints currently accept two possible types,
"l1_norm" and "l2_norm". In both cases, the
norm for each weight vector (including the bias) incident on a neuron
will be constrained to be less or equal than 1. Using data scaled to the
interval, the l1-norm is the one that guarantees best results.
Also, it is important to note that the weight constraints are imposed on all layers except for the last one, which is expected to be linear and not need a Taylor expansion.
Note: Our implementation differs slightly from the constraints provided by
kerasbecause in our implementation we join the bias with the rest of the weights whilekerasdefault constraints allow only for bias or kernel constraints. Our implementation uses a custom callback that is applied at the end of each batch.
nn_uncon <- keras_model()
nn_con <- keras_model()
nn_con <- add_constraints(nn_con, constraint_type = "l1_norm")Training
Now we can compile and train both NNs using standard the
keras approach. Note that we have to increase the number of
needed epochs with the constrained NN to learn properly.
compile(nn_uncon,
loss = "mse",
optimizer = optimizer_adam(),
metrics = "mse")
history1 <- fit(nn_uncon,
train_x,
train_y,
verbose = 0,
epochs = 150,
batch_size = 50,
validation_split = 0.2
)
plot(history1)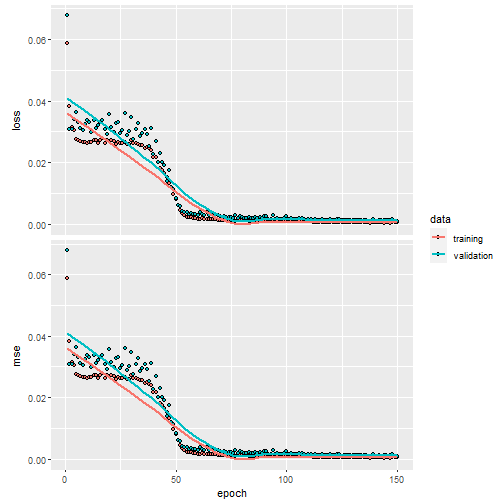
compile(nn_con,
loss = "mse",
optimizer = optimizer_adam(),
metrics = "mse")
history2 <- fit(nn_con,
train_x,
train_y,
verbose = 0,
epochs = 700,
batch_size = 50,
validation_split = 0.2
)
plot(history2)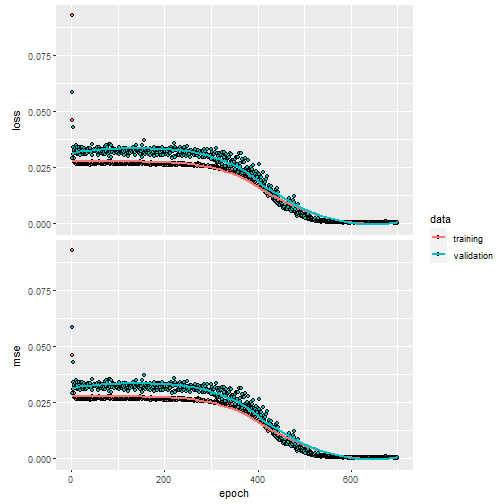
NN predictions
We can visualize the NN predictions vs the original Y values for both neural networks and observe how both of them provide accurate predictions (the values fall near the “perfect” diagonal red line).
# Obtain the predicted values with the NN to compare them
prediction_nn_uncon <- predict(nn_uncon, test_x)
#> 4/4 - 0s - 48ms/epoch - 12ms/step
# Diagonal plot implemented in the package to quickly visualize and compare predictions
nn2poly:::plot_diagonal(x_axis = prediction_nn_uncon, y_axis = test_y, xlab = "Unconstrained NN prediction", ylab = "Original Y")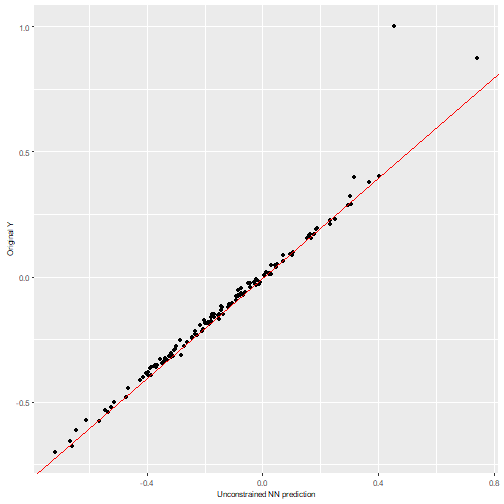
# Obtain the predicted values with the NN to compare them
prediction_nn_con <- predict(nn_con, test_x)
#> 4/4 - 0s - 91ms/epoch - 23ms/step
# Diagonal plot implemented in the package to quickly visualize and compare predictions
nn2poly:::plot_diagonal(x_axis = prediction_nn_con, y_axis = test_y, xlab = "Constrained NN prediction", ylab = "Original Y")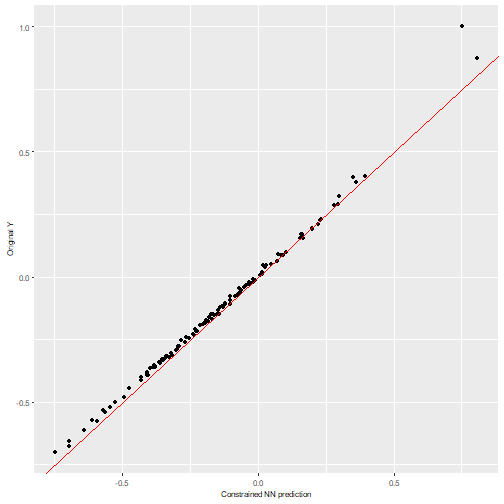
Using nn2poly
After the NNs have been trained, we can directly call
nn2poly on the keras model.
# Polynomial for nn_uncon
final_poly_uncon <- nn2poly(object = nn_uncon,
max_order = 3)
# Polynomial for nn_con
final_poly_con <- nn2poly(object = nn_con,
max_order = 3)We can visualize the polynomial predictions versus NN predictions
# Obtain the predicted values for the test data with our two polynomials
prediction_poly_uncon <- predict(object = final_poly_uncon, newdata = test_x)
prediction_poly_con <- predict(object = final_poly_con, newdata = test_x)
nn2poly:::plot_diagonal(x_axis = prediction_nn_uncon, y_axis = prediction_poly_uncon, xlab = "NN prediction", ylab = "Polynomial prediction") + ggplot2::ggtitle("Polynomial for nn_uncon")
nn2poly:::plot_diagonal(x_axis = prediction_nn_con, y_axis = prediction_poly_con, xlab = "NN prediction", ylab = "Polynomial prediction") + ggplot2::ggtitle("Polynomial for nn_con")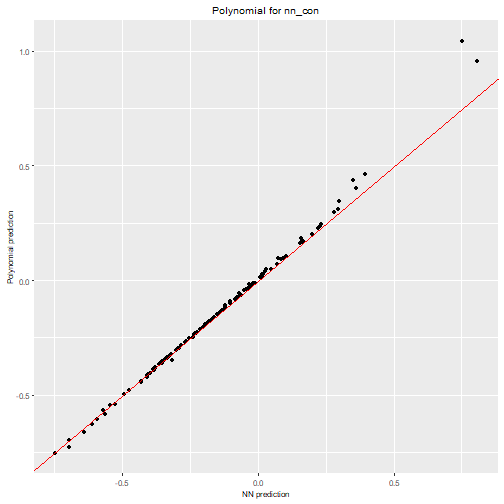
We can clearly see how the constrained NN allows to obtain a close
approximation with nn2poly while the polynomial obtained
for the unconstrained one is not a good representation.
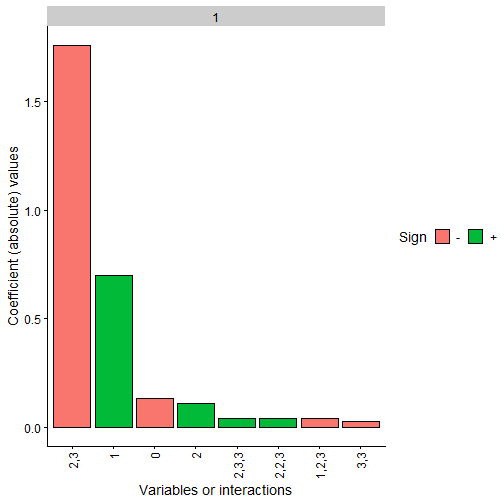
We can also represent the obtained polynomial coefficients and
observe how the constrained case clearly has terms 2,3 and
1 with the same sign as the original polynomial while the
rest are close to 0.
Note: The coefficients values are not in the same scale as the original polynomial due to the fact that we have scaled all the data before training, even the response variable Y. Furthermore, as data has been scaled to the interval, interactions of order 2 or higher would usually need a higher absolute value than the lower order coefficients to be more relevant
plot(final_poly_uncon, n = 8)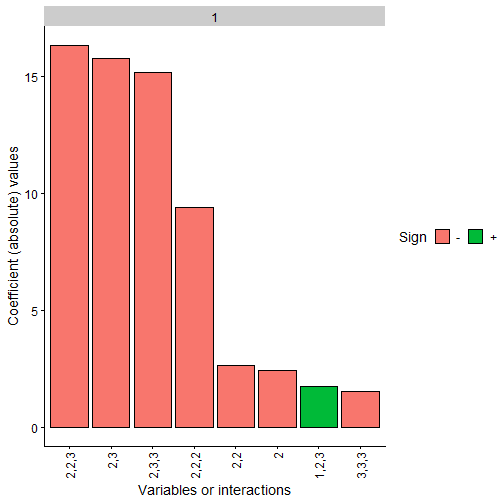
plot(final_poly_con, n = 8)
luz/torch
In this section we will show how to use nn2poly with a
torch neural network, built with its higher level API
luz, and how to impose the needed weight constraints during
training. Furthermore, we will explain how to use
luz_model_sequential(), a helper needed to create a
torch model in an adequate manner so that it can be easily
recognized by nn2poly.
Data loader for torch
In this framework, we need to manipulate a bit our data to use it as
input for a torch model. This can be done dividing our
train_x data intro only train and validation matrices and
using luz::as_dataloader()
# Divide in only train and validation
all_indices <- 1:nrow(train_x)
only_train_indices <- sample(all_indices, size = round(nrow(train_x)) * 0.8)
val_indices <- setdiff(all_indices, only_train_indices)
# Create lists with x and y values to feed luz::as_dataloader()
only_train_x <- as.matrix(train_x[only_train_indices,])
only_train_y <- as.matrix(train_y[only_train_indices,])
val_x <- as.matrix(train_x[val_indices,])
val_y <- as.matrix(train_y[val_indices,])
only_train_list <- list(x = only_train_x, y = only_train_y)
val_list <- list(x = val_x, y = val_y)
torch_data <- list(
train = luz::as_dataloader(only_train_list, batch_size = 50, shuffle = TRUE),
valid = luz::as_dataloader(val_list, batch_size = 50)
)Model definition
Now we will set the structure of our neural networks using a
sequential model in torch. To do so we have implemented the
helper luz_model_sequential(), which lets the user stack
linear layers in a similar way as keras and allows
nn2poly() to be directly used in the model without the need
to extract weights and activation functions manually as in the default
example of vignette("nn2poly-01-introduction").
This following function will be used to create both the constrained
and unconstrained neural networks in the torch example.
luz_nn <- function() {
torch::torch_manual_seed(42)
luz_model_sequential(
torch::nn_linear(p,100),
torch::nn_tanh(),
torch::nn_linear(100,100),
torch::nn_tanh(),
torch::nn_linear(100,100),
torch::nn_tanh(),
torch::nn_linear(100,1)
)
}Now we define both NNs. In this case, differing from the
keras example, we will impose the constraints later to
follow the same approach as in luz documentation. However,
the use of add_constraints() is analogous.
nn_uncon <- luz_nn()
nn_con <- luz_nn()Training
First we train the unconstrained NN using ans standard
luz approach.
fitted_uncon <- nn_uncon %>%
luz::setup(
loss = torch::nn_mse_loss(),
optimizer = torch::optim_adam,
metrics = list(
luz::luz_metric_mse()
)
) %>%
luz::fit(torch_data$train, epochs = 50, valid_data = torch_data$valid)
fitted_uncon %>% plot()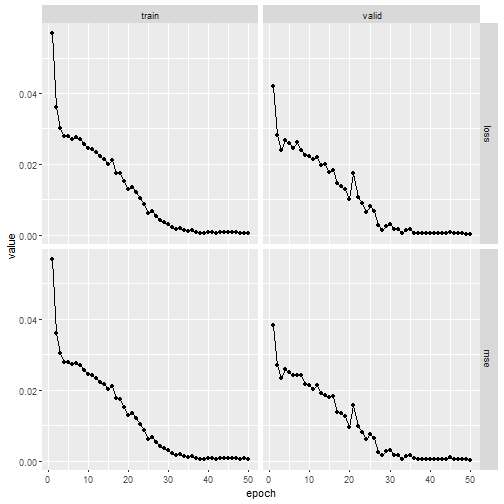
Then, we train the constrained NN by using the function function
add_constraints() before calling fit(). The
constraints currently accept two possible types, "l1_norm"
and "l2_norm". In both cases, the norm for each weight
vector (including the bias) incident on a neuron will be constrained to
be less or equal than 1. Using data scaled to the
interval, the l1-norm is the one that guarantees best results.
Also, it is important to note that the weight constraints are imposed on all layers except for the last one, which is expected to be linear and not need a Taylor expansion.
Note that we have to increase the number of needed epochs with the constrained NN to learn properly.
fitted_con <- nn_con %>%
luz::setup(
loss = torch::nn_mse_loss(),
optimizer = torch::optim_adam,
) %>%
add_constraints("l1_norm") %>%
fit(torch_data$train, epochs = 700, valid_data = torch_data$valid)
fitted_con %>% plot()
Note: Our implementation uses a custom callback that is applied at the end of each batch.
NN predictions
We can visualize the NN predictions vs the original Y values for both neural networks and observe how both of them provide accurate predictions (the values fall near the “perfect” diagonal red line).
# Obtain the predicted values with the NN to compare them
prediction_NN_uncon <- as.array(predict(fitted_uncon, test_x))
# Diagonal plot implemented in the package to quickly visualize and compare predictions
nn2poly:::plot_diagonal(x_axis = prediction_NN_uncon, y_axis = test_y, xlab = "Unconstrained NN prediction", ylab = "Original Y")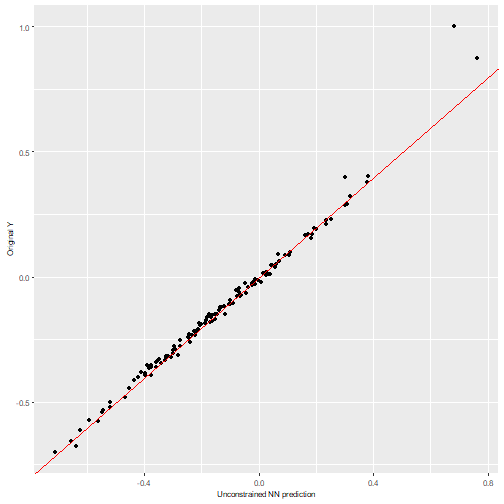
# Obtain the predicted values with the NN to compare them
prediction_NN_con <- as.array(predict(fitted_con, test_x))
# Diagonal plot implemented in the package to quickly visualize and compare predictions
nn2poly:::plot_diagonal(x_axis = prediction_NN_con, y_axis = test_y, xlab = "Constrained NN prediction", ylab = "Original Y")
Using nn2poly
After the NNs have been trained, we can directly call
nn2poly on the luz model.
# Polynomial for nn_uncon
final_poly_uncon <- nn2poly(object = fitted_uncon,
max_order = 3)
# Polynomial for nn_con
final_poly_con <- nn2poly(object = fitted_con,
max_order = 3)We can visualize the polynomial predictions versus NN predictions
# Obtain the predicted values for the test data with our two polynomials
prediction_poly_uncon <- predict(object = final_poly_uncon, newdata = test_x)
prediction_poly_con <- predict(object = final_poly_con, newdata = test_x)
nn2poly:::plot_diagonal(x_axis = prediction_nn_uncon, y_axis = prediction_poly_uncon, xlab = "NN prediction", ylab = "Polynomial prediction") + ggplot2::ggtitle("Polynomial for nn_uncon")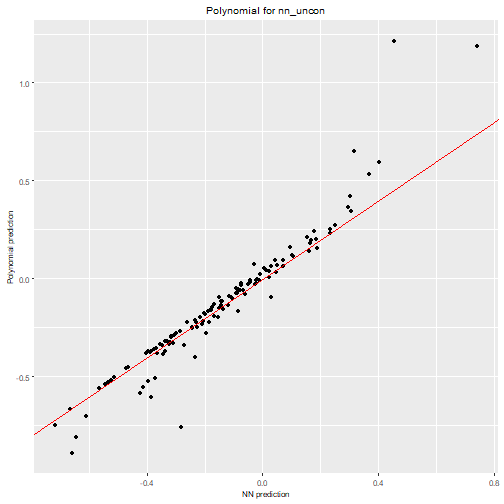
nn2poly:::plot_diagonal(x_axis = prediction_nn_con, y_axis = prediction_poly_con, xlab = "NN prediction", ylab = "Polynomial prediction") + ggplot2::ggtitle("Polynomial for nn_con")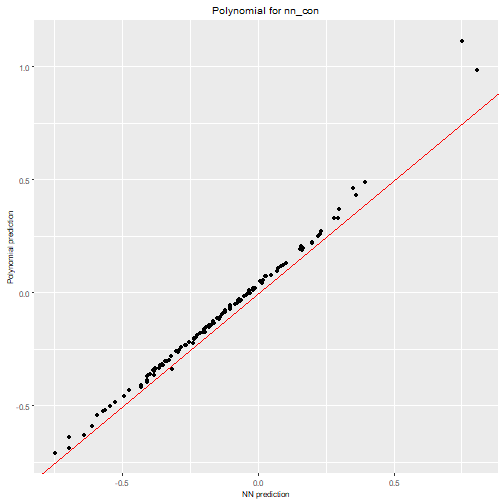
We can clearly see how the constrained NN allows to obtain a close
approximation with nn2poly while the polynomial obtained
for the unconstrained one is not a good representation.
We can also represent the obtained polynomial coefficients and
observe how the constrained case clearly has terms 2,3 and
1 with the same sign as the original polynomial while the
rest are close to 0.
Note: The coefficients values are not in the same scale as the original polynomial due to the fact that we have scaled all the data before training, even the response variable Y. Furthermore, as data has been scaled to the interval, interactions of order 2 or higher would usually need a higher absolute value than the lower order coefficients to be more relevant
plot(final_poly_uncon, n = 8)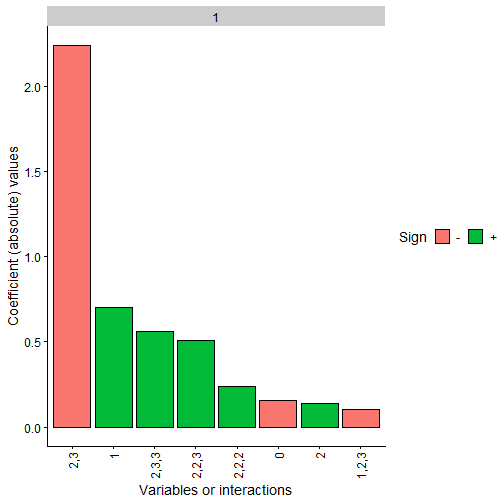
plot(final_poly_con, n = 8)